-Search query
-Search result
Showing 1 - 50 of 308 items for (author: li & yy)

EMDB-35878: 
Cryo-EM structure of DIP-2I8I polymorph 2
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35816: 
Cryo-EM structure of DIP-2I8I fibril polymorph1
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-41277: 
Cryo-EM structure of a SUR1/Kir6.2-Q52R ATP-sensitive potassium channel in the presence of PIP2 in the open conformation
Method: single particle / : Driggers CM, Shyng SL

EMDB-41278: 
Cryo-EM structure of a SUR1/Kir6.2-Q52R ATP-sensitive potassium channel in the presence of PIP2 in the open conformation
Method: single particle / : Driggers CM, Shyng SL

EMDB-43766: 
Kir6.2-Q52R/SUR1 apo closed channel
Method: single particle / : Driggers CM, Shyng SL

EMDB-40799: 
Cryo-EM consensus map of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex
Method: single particle / : Afsar M, Jia L, Ruben EA, Olsen SK

EMDB-35714: 
Cryo-EM structure of Long-wave-sensitive opsin 1
Method: single particle / : Peng Q, Cheng XY, Li J, Lu QY, Li YY, Zhang J

EMDB-35519: 
Cryo-EM structure of hnRAC1-8I fibril
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35520: 
Cryo-EM structure of hnRAC1-2I8I fibril.
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35508: 
Cryo-EM structure of hnRAC1-2I fibril.
Method: helical / : Li DN, Ma YY, Li D, Dai B, Liu C

EMDB-35393: 
Human SLC26A3 in the apo state
Method: single particle / : Li XR, Chi XM, Zhang YY, Zhou Q

EMDB-35264: 
S-ECD (Omicron BA.2.75) in complex with PD of ACE2
Method: single particle / : Li YN, Shen YP, Zhang YY, Yan RH

EMDB-35266: 
S-ECD (Omicron BF.7) in complex with PD of ACE2
Method: single particle / : Li YN, Shen YP, Zhang YY, Yan RH

EMDB-35267: 
S-ECD (Omicron XBB.1) in complex with PD of ACE2
Method: single particle / : Li YN, Shen YP, Zhang YY, Yan RH

EMDB-35268: 
S-RBD(Omicron BA.3) in complex with PD of ACE2
Method: single particle / : Li YN, Shen YP, Zhang YY, Yan RH

EMDB-35269: 
S-RBD (Omicron BA.2.75) in complex with PD of ACE2
Method: single particle / : Li YN, Shen YP, Zhang YY, Yan RH

EMDB-35270: 
S-RBD (Omicron BF.7) in complex with PD of ACE2
Method: single particle / : Li YN, Shen YP, Zhang YY, Yan RH

EMDB-35272: 
S-RBD (Omicron XBB.1) in complex with PD of ACE2
Method: single particle / : Li YN, Shen YP, Zhang YY, Yan RH

EMDB-34929: 
Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form
Method: single particle / : Xu Y, Wu Y, Zhang Y, Fan R, Yang Y, Li D, Yang B, Zhang Z, Dong C, Zhang X, Tang X, Dong H

EMDB-38074: 
CryoEM structure of the histamine H1 receptor in apo-form
Method: single particle / : Wang DD, Guo Q, Tao YY

EMDB-38075: 
CryoEM structure of the histamine H1 receptor-BRIL/Anti BRIL Fab complex with astemizole
Method: single particle / : Wang DD, Guo Q, Tao YY

EMDB-42480: 
Cryo-EM reconstruction of Staphylococcus aureus Oleate hydratase (OhyA) dimer with an ordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

EMDB-42484: 
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer with a disordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

PDB-8ur3: 
Cryo-EM reconstruction of Staphylococcus aureus Oleate hydratase (OhyA) dimer with an ordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

PDB-8ur6: 
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer with a disordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

EMDB-35051: 
D5 ATP-ADP-Apo-ssDNA IS1
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-35052: 
D5 ATP-ADP-Apo-ssDNA IS2
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-35053: 
Cryo-EM Structure of D5 Apo
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-35054: 
Cryo-EM Structure of D5 ADP form
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-35055: 
Cryo-EM Structure of D5 ATP-ADP form
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-35056: 
Cryo-EM Structure of D5 ADP-ssDNA form
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-35057: 
D5 ATPrS-ADP-ssDNA form
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-35058: 
Cryo-EM Structure of D5 Apo-ssDNA form
Method: single particle / : Li YN, Zhu J, Guo YY, Yan RH

EMDB-42502: 
Cyanobacterial RNA polymerase elongation complex with NusG and CTP
Method: single particle / : Qayyum MZ, Imashimizu M, Leanca M, Vishwakarma RK, Bradley Riaz A, Yuzenkova Y, Murakami KS

PDB-8urw: 
Cyanobacterial RNA polymerase elongation complex with NusG and CTP
Method: single particle / : Qayyum MZ, Imashimizu M, Leanca M, Vishwakarma RK, Bradley Riaz A, Yuzenkova Y, Murakami KS

EMDB-34927: 
Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram
Method: single particle / : Xu Y, Wu Y, Zhang Y, Fan R, Yang Y, Li D, Yang B, Zhang Z, Dong C, Zhang X, Tang X, Dong H
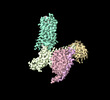
EMDB-35297: 
Structural insights into human brain gut peptide cholecystokinin receptors
Method: single particle / : Ding Y, Zhang H, Liao Y, Chen L, Ji S

EMDB-17435: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion in complex with Arf3
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

EMDB-17436: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

EMDB-17437: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin, the central region of the toxin
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

EMDB-17438: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin, the tail region of the toxin
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

EMDB-17439: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin, the head of the toxin
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

EMDB-17440: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

EMDB-17450: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin, the low-resolution consensus map
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

PDB-8p50: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion in complex with Arf3
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

PDB-8p51: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S

PDB-8p52: 
Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin
Method: single particle / : Belyy A, Heilen P, Hofnagel O, Raunser S
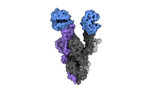
EMDB-40976: 
Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B
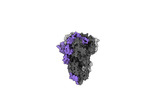
EMDB-40977: 
Cryo-EM structure of mink variant Y453F trimeric spike protein
Method: single particle / : Ahn HM, Calderon B, Fan X, Gao Y, Horgan N, Zhou B, Liang B
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
